Autophagy overall solution
Autophagy or autophagocytosis: also known as type II cell death, is a process in which cells degrade their damaged organelles and macromolecules by using lysosomes under the regulation of autophagy related gene (ATG). Autophagy, as a research hotspot, has a very far-reaching research significance. Topbiotech has rich experience in autophagy research. We can provide you with an overall solution from subject design, experimental operation, experimental results and data analysis.
Autophagy inducer
1)Bredeldin A / Thapsigargin / Tunicamycin :Simulated endoplasmic reticulum stress;
2)Carbamazepine/ L-690,330/ Lithium Chloride:IMPase inhibitor (Inositol monophosphatase);
3)Earle's balanced salt solution (BSS): manufacturing hunger;
4)N-Acetyl-D-sphingosine(C2-ceramide):Class I PI3K Pathway inhibitor;
5)Rapamycin:mTOR inhibitor(commonly used);
6)Xestospongin B/C:IP3R blocker.
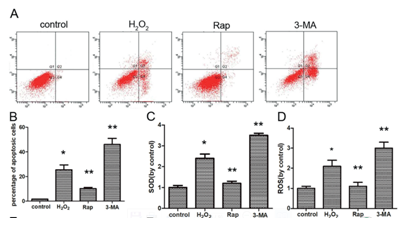
Autophagy inhibitor
1)3-Methyladenine (3-MA): (Class III PI3K) hVps34 inhibitor;
2)Bafilomycin A1: Proton pump inhibitor;
3)Hydroxychloroquine.
Detection of morphological characteristics of autophagy
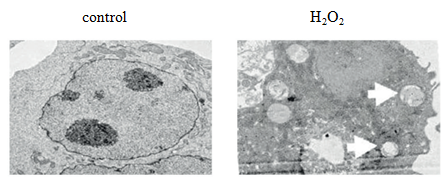
1) Autophagosomes are observed by transmission electron microscopy.
Autophagosome is a landmark structure of autophagy. Autophagosome belongs to subcellular structure, with a diameter of 300 ~ 900 nm and an average of 500 nm, which can not be seen under light microscope. Through transmission electron microscopy, autophagosome has always been the most direct and classic method to observe autophagy, and it is the "gold standard" of autophagy detection.
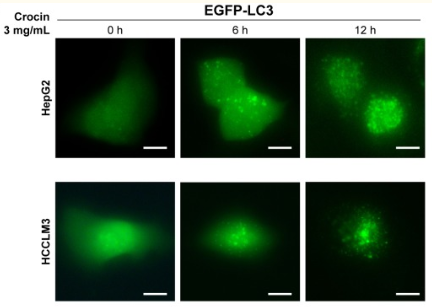
2) GFP-LC3 fusion protein was used to trace autophagy formation under fluorescence microscope.
Since LC3 is required during autophagy membrane formation, autophagy can be recognized by labeling LC3.
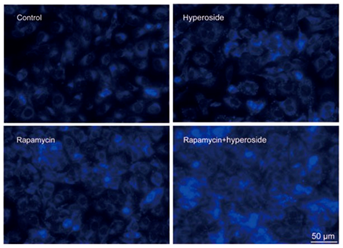
3) Fluorescence staining
Tissue sections or cells were stained with fluorescent dyes to observe the occurrence of autophagy. According to this characteristic, autophagy lysosomes can be labeled with acid oriented dyes MDC, acridine orange and lyso tracker.
Autophagy marker protein
1)Autophagy marker protein LC3
Microtubule associated protein 1 light chain 3 (LC3 / Atg8) is a marker protein on autophagy membrane. There are two forms of LC3 protein in cells, LC3 - Ⅰ and LC3 - Ⅱ. The C-terminal of newly synthesized LC3 in cells was cleaved by atg4 protease to form cytoplasmic soluble LC3 - Ⅰ. When autophagosomes are formed, LC3 - Ⅰ is modified by shearing and ubiquitination. LC3 - Ⅱ is coupled with phosphatidylethanolamine (PE) on the surface of autophagy membrane to form a membrane-bound form and is located in the inner and outer membrane of autophagy. Therefore, the ratio of LC3 - Ⅱ / Ⅰ can estimate the level of autophagy
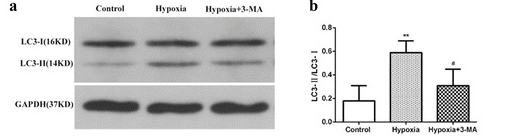
2) Detection of autophagy substrate protein
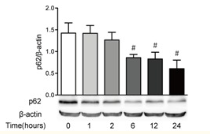
p62 is coupled to LC3 and participates in the composition of autophagosomes as a regulatory factor. It is degraded in the middle and late stages of autophagy. There is a negative correlation between the expression of p62 and autophagy activity. The decrease of p62 level detected by Western blot can reflect the degree of autophagy activity. The increase of p62 implies that autophagy and lysosomal degradation pathway are inhibited.
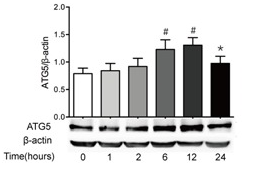
3) Atg12-atg5 complex
In addition to LC3, there are atg12-atg5 complexes on autophagy membrane. Atg12 and ATG5 are covalently bound together like a single molecule after translation, and it is located in the whole extension stage of autophagy bilayer isolation membrane.
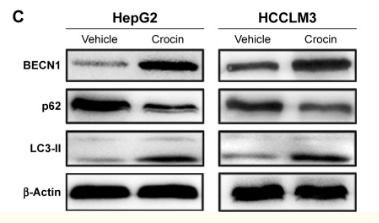
4)Beclin1
Mammalian Beclin1 is a homologue of yeast atg6 gene. Beclin1 controls cell growth and tumor inhibition mechanisms by activating autophagy. Western blot detection of Beclin1 protein expression directly reflects autophagy activity.
5) Autophagy related marker proteins also include Atg1, Atg9, dram1, atg14, atg16 and atg18.
Regulatory pathway of autophagy
1) Autophagy dependent on mTOR (mammalian rapamycin target) pathway
A.PI3K-AKT-mTOR signal pathway

B. AMPK-TSC 1/2-mTOR signal pathway
2) Other signal pathways
A. 3-methyladenine (3-mA) inhibits autophagy by inhibiting the activity of class Ⅲ PI3K;
B. Beclin1 and UVRAG as positive regulators and anti apoptotic factor Bcl-2 as negative regulators participate in the formation of class Ⅲ PI3 complex to regulate autophagy;
C. GTP bound G protein subunit Gαi3 inhibits autophagy; GDP combined Gαi3 protein activated autophagy;
D. Autophagy was induced by death associated protein kinase (DAPK) and DAPK related protein kinase-1 (Drp-1).

